CRISPR technology has contributed in recent years to the revolution of genetic engineering, allowing genes to be modified in a simple way. This system is composed of 2 key elements: the RNA guide that binds to the target DNA, to direct the second component, Cas9, which cuts target DNA by recognizing a sequence called PAM.
It is quite a challenge to choose the components of the CRISPR system. In a previous post, we pointed out some tips for successfully designing RNA guides. It may seem a complicated task, but by following a few tips and taking the advantage of online tools, success is almost guaranteed.
In this post we will touch base on how to minimize and detect off-target effects. Off-target effects occur when the guide binds to other parts of the target DNA, different from the sequence which it was designed for. This action causes CAS9 to cut the DNA at other points, generating the introduction of other mutations not expected. It is obvious that a good RNA guides design is essential to minimize off-target effects.
Therefore, the detection of these unwanted effects is necessary. Although CRISPR technology has evolved and grown exponentially in recent years, it is still not fully understood. Thus, there are still many aspects to investigate and discover.
Abyntek makes this revolutionary technology accessible to you accompanying you throughout the entire process.
The off-target detection task must be carried out in two steps:
1) Prediction of possible in silico effects, using computational tools. They are known as biased tools since the information they give is predictions that may or not will be fulfilled.
2) Subsequent experimentation using unbiased methods. This makes possible to detect the cuts outside the target, throughout the whole genome.
Web-based tools
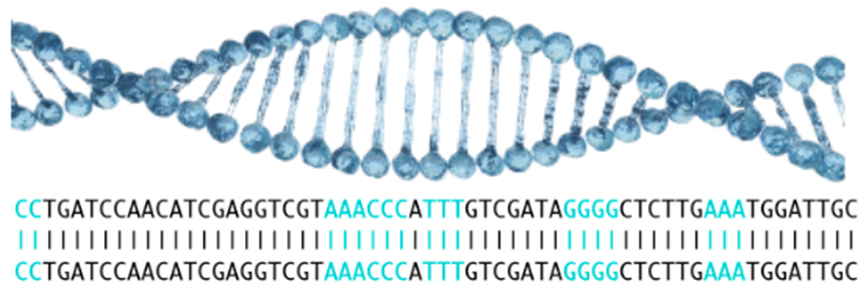
Traditional tools based on alignment
At the beginning, it was thought that the existing traditional alignment tools (short sequence alignment) could be used for CRISPR. Thus, the target site was realigned with the reference genome in order to identify similar sites that might be inadvertently targeted by CAS9. although BLAST has been used in its origins, BWA or Bowtie work better allowing this type of alignment.
However, these refurbished or upgraded tools are not a solution for use in CRISPR experiments. The search for potential off-target sites requires the identification of small sequences corresponding to 20 bp + the PAM sequence, which can lead to certain mismatches. These traditional tools are not prepared to detect such small and divergent sequences. Bowtie and BWA allow 3 and 5 mismatches respectively, which may mislead some off-targets.
Comparing traditional methods versus those designed specifically for the CRISPR-CAS9 system, the first ones are not considered specific enough to be used for CRISPR experiments. Traditional methods not only might miss off-targets with large mismatches, but also some with just one.
Alignment based CRISPR tools
To accurately identify potential off-targets, new alignment methods have been implemented, specifically created for the CRISPR system. In this type of method, two bidirectional aligners first match a small portion of the sequence, called the seed. Each aligner works by extending this initial seed region in both directions. This way of working simultaneously allows all possible off-targets to be identified.
It must be taken into account that not all the presumed off-targets are functional, so false positives can be generated. To reduce the number of false positive predictions, these predictive models limit potential off-targets by maximizing the number of mismatches. However, it has also been observed that the real off-targets may be different from those predicted, resulting in false negatives. Therefore, a balance of false positives and negatives is necessary. To compensate for this, classification algorithms have been developed, being able to filter out false positives.
Some of the most used programs in CRISPR experiments are CRISPOR or Cas-OFFinder. They incorporate all these algorithms and advances to predict off-target effects. Cas-OFFinder is considered the universal off-target detection tool.
However, although these tools are very useful, as already mentioned, they are based on predictions. Thus, the most recommended way to proceed when designing a CRISPR experiment is to predict possible off-targets in silico, and then confirm predictions in vitro or in vivo.
In vitro tools
Whole genome sequencing (WGS) is likely one of the best approaches to verify off-target cuts. This technique allows detecting and quantifying the introduced mutation, and additionally, whether other off-target areas have mutated.
However, this method also has drawbacks. For that reason, improved techniques based on this methodology have been developed, such as digenome-seq, site-seq, or circle-seq. These techniques, like all of them, have their advantages and disadvantages, as well as recommendations depending on the application. Digenome-seq and site-seq are based on the digestion of the cell genome with the ribonucleoprotein complex (RNP), thus maximizing the detection of off-targets that are later detected by massive sequencing (WGS) as fragments with 5´ identical endings.
On the other hand, circle-seq was developed to reduce the background noise of previous techniques. To do this, the DNA is circularized first, and then linearized again. The next step is linear DNA sequencing, which virtually eliminates detection of previous DBS.
In vivo tools
In vivo tools have been designed for some types of experiments, where in vitro off-targets do not represent what happens in vivo.
The first tool developed for this purpose was IDVL, based on IDLV lentiviral vectors (integrase defective lentiviral vectors). The lentiviral vector is transferred simultaneously, together with the CRISPR system, which is integrated upon mutation correction by DBS. Subsequently, the DNA is cut and fragments are religated using a linker. To determine the off-target effect, a PCR is performed to detect the distribution of IDLVs. This system is very effective in cells that are difficult to transfect, such as primary cells.
Based on this strategy, other methods have been developed, such as BLESS or GUIDE-Seq, which try to overcome the arisen limitations. One of the most robust and sensitive in vivo techniques is LAM-HTGTS. This approach allows the detection of chromosomal translocations in mammalian cell cultures, through the binding of DSBs produced by endonucleases. However, translocations due to DSBs are rare.
There are other in vivo techniques with their advantages and disadvantages, such as DISCOVER-Seq, VIVO, or the latest to be developed, GOTI. The latter allows detection of off-targets in a cell population from a single blastomere. In this way, off-target sites can be detected in a mouse embryo in the early stages of development.
All the methodologies described here are just some examples available. They all have their advantages, disadvantages and recommended applications. CRISPR knowledge increase daily. This, allows new techniques and approaches to emerge, improving previous limitations.
At Abyntek, we provide an extensive catalogue and custom products validated by The Broad Institute of MIT and Harvard ensuring the best quality service. Our experienced scientific team will guide you, boosting your research to another level.
Tips for minimizing off-target effects
Guide design
Computational tools designed for CRISPR, such as CHOPCHOP and CRISPOR, take the following rules into account when designing RNA guides:
– The position of missmatches affects off-target effects.
– The potential off-targets must not be followed by the PAM 5’NGG or 5’NAG sequences. Missmatches close to PAM sequences are less well tolerated.
– The minimum number of mismatches between the sgRNA and the off-target potentials must be limited to 3 nucleotides.
– Mismatches at the 5′ end of the sgRNA are better tolerated in the first 8-14 base pairs.
– Off-targets can occur even with 6 mismatches between gRNA and off-target DNA.
– Adjusting the Cas9:gRNA ratio improves the apparition of off-targets (Hsu 2013).
– Use truncated sgRNA
Delivery systems

The introduction of the CRISPR system into the cell through RNP reduces off-target effects compared to plasmids.
This system uses lipofectamine and charged nanoparticles as a delivery system. Before entering into the cell, it binds to DNA and directly cuts it. Subsequently, it is degraded very quickly, reducing the exposure time, and thus the probability of cutting out of the target. In contrast, in plasmid systems, the RNA has to be transported into the nucleus and then the mRNA translocated out for translation. The expressed Cas9 and gRNA then have to be returned to the nucleus in order to edit the target DNA. This process usually takes about 24 hours. Furthermore, plasmids are capable of surviving days inside the cell. This increases the probability of producing more off-targets.
Cas enzyme election

The most commonly used nicase enzyme in CRISPR experiments is Streptococcus pyogenes Cas9 (spCas9). However, it has been modified to try to reduce off-target effects. One of this modified Cas9 is eSpCas9 (enhanced specificity). Some of the amino acids of this enzyme do not make specific contact with the target DNA, so they are mutated to reduce the number of tolerable mismatches. This revamped eSpCas9 was named SpCas9-HF1 or high-fidelity. On the other hand, although these two new nicases are more specific, the effect on the target DNA (on-target) was also reduced in some genes.
New improvements have been added to this enzyme, until create the last one, called HiFi Cas9. This one has a point mutation that increases activity at the target site, maintaining a very low, practically undetectable, off-target activity.
There are also Cas enzymes from other microorganisms, with different characteristics. For example, SaCas9, from S. aureus. This nicase is encoded by a smaller gene, allowing its use with with an adeno-associated vector (AVV), for more efficient in or ex vivo applications. Furthermore, it is capable of recognizing longer PAM sequences, which likely increases specificity.
Do you need any help? Contact us filling this form or sending an email to info@abyntek.com. We will be pleased to assist you!
Recommended bibliography for more information about these techniques:
Doi: 10.3389/fgeed.2021.673022
Doi: 10.3390/cells9071608
Doi: 10.3389/fcell.2021.718466




