CRISPR technology has become a game-changer in the field of genome editing. In the last decade, the numbers of applications have increased exponentially and labs from all around the word are making use of this technology to address their scientific questions.
If you want to know more about where does this technology come from, how does the system work, its applications and much more, have a look at our new post!
1. What is CRISPR and where does it come from?
CRISPR stands for Clustered Interspaced Short Palindromic Repeats.
It refers to repetitive DNA sequences with ´spacer´ DNA sequence that were observed in bacteria and match viral sequences. It was later discovered that it constitutes the adaptive immune system of prokaryotes: they integrate short virus sequences in the CRISPR locus so that these elements can be transcribed upon viral infection to target the virus by cleaving their nucleic acids. This defence mechanism also requires the presence of CRISPR-associated (Cas) genes, which code for proteins that are essential of the immune response, thus it is referred to as CRISPR-Cas system. Different CRISPR-Cas systems have been described for different bacteria and archaea, and they differ in their components as well as in their mechanism of action. In the last decade, a variety of tools based on the CRISPR-Cas systems have been developed and are being used in genome editing with great specificity.
2. How does the CRISPR-Cas9 system work?
The first CRISPR system that was engineered for its application in genome editing for mammalian cells is that of Streptococcus Pyogenes. It comprises the nuclease Cas9 which can generate a blunt double strand break (DSB) following DNA target recognition. To be guided to the specific sites of the genome, Cas9 forms a complex with a CRISPR RNA (crRNA) and a pair of trans-activating CRISPR RNAs (tracrRNAs) that facilitate ribonucleoprotein complex formation. These RNA molecules have been engineered into a single guide RNA (sgRNA) to facilitate genome editing applications. The sgRNA is designed to contain a 20-nucleotide long sequence that pairs with the DNA target sequence. Of note, the CRISPR-Cas systems also require a short sequence, known as the protospacer-adjacent motif (PAM) to be present near the target DNA site. For the CRISPR-Cas9 system the PAM sequence (5´ to 3´) is NGG, where N represents any nucleotide. Other naturally occurring CRISPR systems rely on additional Cas9 orthologues that can recognize different PAM sequences thereby expanding the choice of target sites. Upon DNA target recognition Cas9 cleaves the DNA few base pairs upstream of the PAM sequence. The DSB that is generated will trigger a DNA damage response to stimulate repair by various mechanisms that can be harnessed for genome editing.
3. What are the CRISPR-Cas9 applications in gene editing?
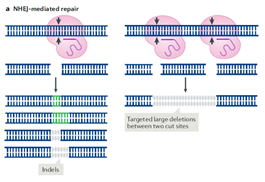
The DSBs generated by nucleases can be repaired by distinct DNA repair mechanisms which can be harnessed for specific gene editing purposes. The two major repair mechanisms utilized are:
- Non-Homologous End Joining (NHEJ)-mediated repair: this mechanism is prone to error and induces some insertions or deletions (indels) after several cycles of break and repair.
- Homology-Directed (HDR)-mediated repair: in the presence of a donor template the DSB can be repaired without errors. However, this repair mechanism occurs at a lower efficiency in comparison to NHEJ, and typically requires the cells to be undergoing division.
Based on the repair mechanism described above, the following precise genomic alterations can be made:
- Gene deletions: to create a gene knockout a common strategy is to target a coding exon at the beginning of your gene of interest. The DSB will undergo NHEJ-mediated repair which very likely will introduce frameshift mutations thereby disrupting gene expression. An alternative strategy consists in simultaneously targeting two sites in a gene to delete the sequence generated between the two DSBs.
- Gene insertions: inserting a DNA sequence into a protein-coding gene can be achieved by providing a donor template. The donor template can be incorporated in the genome via homologous recombination. To accomplish this, the exogenously provided donor template should contain homology regions to the sequences flanking the location of the break. Synthetic single-strand or double strand DNA donor oligos can be delivered into a cell to induce small changes, such as point mutations, within the genomic target region. By the same means, epitope tags can also be introduced.
- Translocations: Translocations can occur when DSBs are generated in two non-homologous chromosomes and these DSBs on the two chromosomes are improperly joined. Cas9-induced chromosomal rearrangements have been introduced to mimic for instance, cancer-relevant translocations or oncogenic gene fusions.
Reference: (Pickar-Oliver & Gersbach, 2019)
4. What is a base editor?
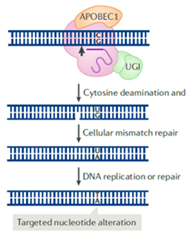
While HDR-mediated repair of DSBs can be used to introduce point mutations, it also has some limitations. Firstly, is not efficient in post-mitotic cells and secondly, the introduction of DSBs might result in off-target mutagenesis. To overcome this, tools have been developed based on a Cas9 nickase (Cas9n), which is a variant of Cas9 engineered so that it only cleaves one of the DNA strands. Fusing Cas9n to a single stranded DNA deaminase enzyme, two main classes of base editors have been developed:
- Cytosine-based editors (CBEs, catalyse the conversion of C/G base pair to T/A base pair)
- Adenine-based editors (ABEs, catalyse the conversion of A/T base pair to G/C base pair).
In both cases the Cas9n localizes the ssDNA deaminase enzyme to a particular target sequence, allowing the introduction of single nucleotide changes without generating DSBs.
Reference: (Pickar-Oliver & Gersbach, 2019)
5. How do I plan my CRISPR experiment? Where do I start?
In the last decade numerous CRISPR-based tools have been developed, giving researchers multiple options to conduct gene editing. Thus, when designing your CRISPR experiment, one of the main things to take into consideration is the type of modification desired. This will determine which are the most suitable CRISPR reagents. Below we describe the steps of things to take into consideration:
- Step 1: Determine your genetic modification. To generate gene knockouts the gRNA should be design to target the 5´ exon or essential protein domains.
- Step 2: Select the expression system: CRISPR-Cas9 components can be delivered in vivo via different systems:
- Plasmid vector: Expression of Cas9 and gRNA. The plasmid can also contain reporter or selection markers.
- Lentiviral vector: For cell types that are difficult to transfect. Expression of Cas9 and gRNA
- AAV Vector: Transient or stable transfection of SaCas9 (smaller than Cas9 to allow packaging into a single AAV vector) and/or gRNA. For infection of dividing and non-dividing cells.
- Cas9 mRNA and gRNA: allows a transient expression of CRIPSR components. The delivery can be made via microinjection or electroporation
- gRNA/Cas9 Ribonucleoprotein complex: allows a transient expression of CRIPSR components. Cas9 protein is purified and gRNA in vitro transcribed.
- Step 3: Select target DNA sequence and design gRNA. There are genome-wide gRNA databases and design tools which we highly recommend to choose the gRNA. These tools also determine or predict the on/off target activity.
- Step 4: For knockins, design the donor construct. HDR donor constructs can be delivered as plasmids or ssDNA oligos. The donor template should be designed with mutations in the PAM sequence to prevent cleavage once it has been incorporated.
- Step 5: Synthesize gRNA/Cas9 vectors. Customized vectors can be cloned or commercially ordered.
Are you thinking about using CRISPR but you are unsure as to where to start? At Abyntek we would be thrilled to hear about your projects and help you out with the strategy and choice of reagents! Don´t hesitate to contact us for more information.
References:
Anzalone, A. V, Koblan, L. W., & Liu, D. R. (2020). Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nature Biotechnology, 38(7), 824–844. https://doi.org/10.1038/s41587-020-0561-9
Pickar-Oliver, A., & Gersbach, C. A. (2019). The next generation of CRISPR-Cas technologies and applications. Nature Reviews. Molecular Cell Biology, 20(8), 490–507. https://doi.org/10.1038/s41580-019-0131-5