There is an increasing body of research work regarding studies of the gastrointestinal microbiota as the cause of, or potential treatment for, various medical conditions. To gain a better understanding of recent progress in this field, herein we present a brief review of the applications of fecal transplantations to treat recurring Clostridium difficile infections, the implications of genetic variants on the composition of the intestinal microbiota, and models for establishing a causal relationship between changes to the intestinal microbiota and various medical conditions.
Fecal transplantation for treating Clostridium difficile infections
C. difficile is the leading cause of infectious diarrhea in a hospital setting. Although some patients may remain asymptomatic after exposure to this pathogen, many develop symptoms ranging from mild diarrhea to the onset of colitis, with the associated risk of death. One of the main risk factors for suffering this disease is the administration of antibiotics, as they provoke changes in the intestinal barrier. Although antibiotics are essential for treating potentially fatal diseases, they also affect our intestinal microbiota, altering its protective function and facilitating the growth of C. difficile. Age (>60 years) and the presence of other comorbidities are risk factors known to increase the mortality rate.
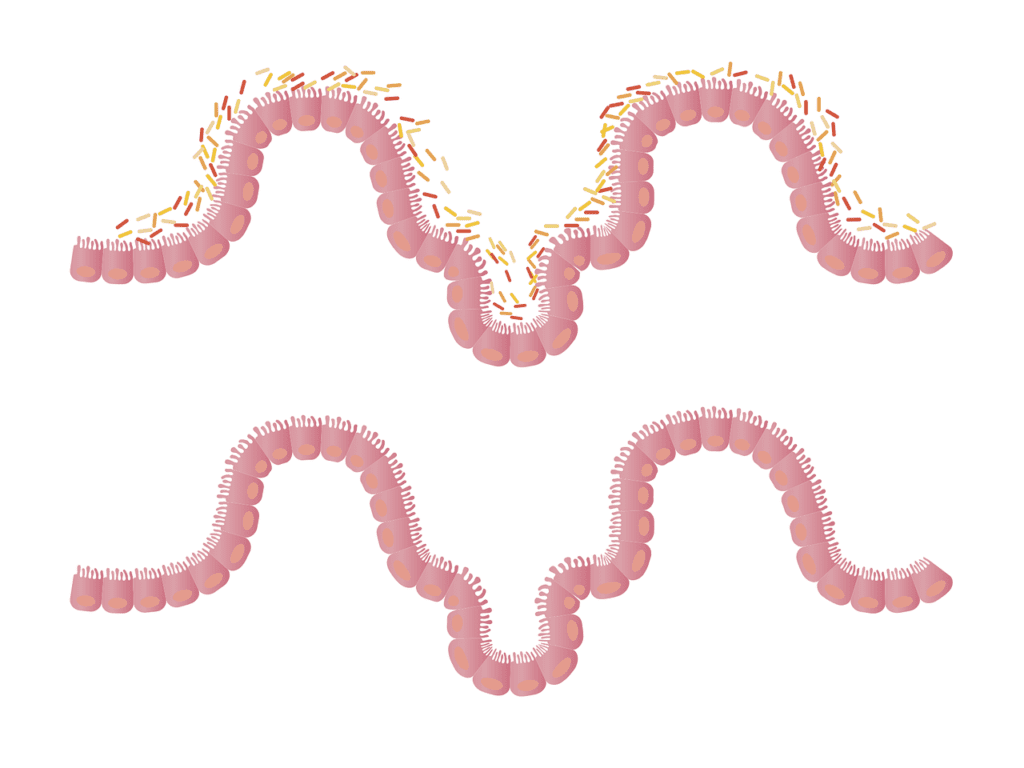
Microbial agents that specifically target C. difficile are used to combat this infection.However, the efficacy of these treatments is limited as they are unable to act against the main mechanism of infection, namely the spores. Spores can be present in the environment or even enter the food chain. One strategy that has allowed recurring C. difficile infections to be treated effectively is known as fecal microbiota transplantation (FMT). This procedure involves applying a fecal solution from a healthy donor to the patient’s intestinal tract in order to re-establish the composition of their intestinal microbiota and hinder the propagation of the pathogen. The high rate of success achieved using this strategy highlight the close relationship between the composition of our intestinal microbiota and our susceptibility to C. difficile infection.
Do genetic factors affect the composition of our intestinal microbiota?
It should be noted that our gastrointestinal microbiota affects more than just C. difficile infection. Indeed, numerous human diseases have been associated with a fecal microbiota that differs from that of healthy individuals in one way or another. In particular, differences are observed in the diversity and relative abundance of some microbes and in the metabolites produced by those species. The term “dysbiosis” has been used to refer to alterations to the microbiota when compare to “normal”, and such alterations have been observed in patients with allergies, Parkinson’s disease, or inflammatory bowel disease, to name just a few examples.
However, what do we considered to be “normal”? Although the microbiota field has come on in leaps and bounds in recent years, the characteristics of a “healthy” or “normal” microbiota have not yet been described in detail. Numerousfactors , such as diet, use of antibiotics, age, or even environmental factors, affect the composition of a person’s gastrointestinal microbiota. Moreover, this variability may also have a genetic cause. In this regard, various studies published over the past few years have shown that some genetic variants or single nucleotide polymorphisms (SNPs) are associated with a specific composition of the gastrointestinal microbiota. However, it is currently difficult to establish solid conclusions regarding these associations as, in the majority of cases, the SNPs identified vary from one study toanother . Despite this, there is one exception that appears to be common to most studies.
- Variants in the gene for the enzyme lactase (Lct) are significantly associated with a higher relative abundance of Bifidobacterium in the intestinal tract. In particular, the presence of the CC genotype in SNP rs4988235 results in a low activity for this enzyme. As Bifidobacterium preferably obtains its energy by metabolizing lactose, it can be speculated that individuals with a high enzyme activity have a lower abundance of Bifidobacterium in their microbiota as the availability of lactose is also lower. In contrast, individuals with a lower enzyme activity have more lactose available, thereby allowing Bifidobacterium to growth.
Nevertheless, it is likely that the genetic associations observed are also affected by environmental factors to some degree. Specifically, the consumption of lactose-containing products may affect the association observed with the Lct gene.Therefore, a more in-depth study of the effects of both these factors (genetics and the environmental) will be necessary to draw firm conclusions.
Establishing a causal relationship between intestinal dysbiosis and disease
A crucial question that remains to be answered is the nature of the relationship between intestinal dysbiosis and disease. Are changes to the microbiota responsible for some diseases or a consequence of them? In light of this, it is essential to establish a cause-effect relationship if we are to consider the gastrointestinal microbiota to be a valid therapeutic target for the treatment of a number of conditions.

The most widely used model for establishing such a causal relationship involves performing a fecal microbiota transplantation from individuals with or without a particulardisease into experimental mice lacking a microbiota, also known asgerm-free mice.By performing a comparative analysis between both groups and monitoring progress of the disease it can be established whether there is the microbiota can induce disease. This model, which is known as Human Microbiota-Associated mice (HMA mice), not only allows a causal relationship to be established, but can also serve as mean to dissect the molecular mechanism leading to disease, and explore potential therapeutic interventions.
Despite its limitations—extrapolation of the results from mice to humans should be done cautiously—this model has shown that Helicobacter pylori is one of the main causes of peptic ulcers and that the microbiota plays a protective role against C. difficile. To date, robust and conclusive studies demonstrating a causal relationship between the intestinal microbiota and other conditions, such as type 2 diabetes, rheumatoid arthritis, or neurological diseases, are still lacking. Nonetheless, some studies have clearly associated these diseases with gastrointestinal dysbiosis. Future studies aiming to address causality will be essential to stablish the basis of a new era of treatments designed to modulate our microbiota as a mean to treat disease.
References:
Blekhman, Ran, Julia K Goodrich, Katherine Huang, Qi Sun, Robert Bukowski, Jordana T Bell, Timothy D Spector, et al. 2015. “Host Genetic Variation Impacts Microbiome Composition across Human Body Sites.” Genome Biology 16 (1): 191. https://doi.org/10.1186/s13059-015-0759-1.
Cahana, Inbal, and Fuad A Iraqi. 2020. “Impact of Host Genetics on Gut Microbiome: Take-Home Lessons from Human and Mouse Studies.” Animal Models and Experimental Medicine 3 (3): 229–36. https://doi.org/10.1002/ame2.12134.
Durack, Juliana, and Susan V Lynch. 2019. “The Gut Microbiome: Relationships with Disease and Opportunities for Therapy.” The Journal of Experimental Medicine 216 (1): 20–40. https://doi.org/10.1084/jem.20180448.
Turpin, Williams, Osvaldo Espin-Garcia, Wei Xu, Mark S Silverberg, David Kevans, Michelle I Smith, David S Guttman, et al. 2016. “Association of Host Genome with Intestinal Microbial Composition in a Large Healthy Cohort.” Nature Genetics 48 (11): 1413–17. https://doi.org/10.1038/ng.3693.
Walter, Jens, Anissa M Armet, B Brett Finlay, and Fergus Shanahan. 2020. “Establishing or Exaggerating Causality for the Gut Microbiome: Lessons from Human Microbiota-Associated Rodents.” Cell 180 (2): 221–32. https://doi.org/10.1016/j.cell.2019.12.025.
If you are interested in collecting stool samples for your own study of the microbiota, further information regarding OMNI-Gene GUT kits can be found here.
Information regarding kits for obtaining DNA from saliva (Oragene DNA) to perform genetic studies is also available here.